All courses funded by this research school will make all of its material freely available online to everyone in the world using the Creative commons 3.0 license. Lectures may be recorded on video and put online.
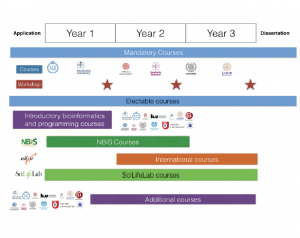
Compulsory courses (3-5 hp)
The basis for this research school is two compulsory courses per year for the students. The courses are progressive, i.e. the learning outcomes from the first course are a necessary requirement for the next course. The earlier courses are aimed at teaching important computational methods, while the later courses are aimed at relevant high-throughput methods for medical bioinformatics.
First-year courses:
1. Applied Bioinformatics, 3 hp (Samuel Flores, Swedish University of Agricultural Sciences. In this course the students will be taught a good working habits for bioinformatics. They will learn to work in Unix, using Python for scientific code development and learn about good practices for scientific programming. Tentative maximum enrollment of 30 students. Includes pre-course assignments. Earlier course editions:
- In-person 28 May – 1 June, 2018 at SciLife Lab, Solna.
- Online 11-12 and 15-17 June, 2020.
- Online 14 -18 June, 2021.
- In-person 13-17 June 2022, at the Swedish University of Agricultural Sciences in Ultuna, Uppsala.
- In-person May 29 – Jun 2, 2023, at the Swedish University of Agricultural Sciences, Campus Ultuna, Uppsala.
- In-person May 27 – 31, 2024, at the Swedish University of Agricultural Sciences, Campus Ultuna, Uppsala.
- In-person October 6 – 10, 2025, at the Swedish University of Agricultural Sciences, Campus Ultuna, Uppsala.
2. Statistics in Bioinformatics, 3HP (Dirk Repsilber, Örebro Univ and Zelmina Lubovac, Skövde Univ). Replaces Algorithms in Bioinformatics, 5HP. After completing the course, the student should understand and by means of statistical bioinformatics methodology have ability to perform: (i) basic data analysis based on univariate and multivariate statistical principles, (ii) omics data analysis, (iii) identification of predictive biosignatures for precision medicine. There are two examination parts in the course: pre-course assignment, and individual project report. Pre-course assignment must be completed before the course start. Earlier course editions:
- Online, 19 to 23 October, 2020.
- In-person, 22-26 November, 2021 at Skövde University, Skövde.
- In-person, November 21 – 25, 2022, at Örebro University.
- In-person, November 20 – 24, 2023, at Örebro University.
- In-person, November 24 – 28, 2025, at Örebro University. A lodging offer have been negotiated, hotel code will be provided
Second-year courses:
3. Machine Learning in Medical Bioinformatics, 5HP (Björn Wallner, Linköping Univ). This workshop will consist of a mix between research talks, the theory behind the research, and actually applying the theory yourself in practical exercises. Topics discussed during the workshop: unsupervised and supervised learning and various machine learning techniques, ranging from linear models, support vector machines, random forests, neural networks, and deep neural networks. Pre-course assignment must be completed before the course start. From 2019 to 2024 the course was given yearly, meeting for one week sometime between 22 april and 21 may at Linköping University.
The 2025 edition will run 5-9 May, with the on-site portion running 6-9 May in Linköping.
4. Experimental design for high-throughput technologies, 3 HP (Carsten Daub, KI). In this course the students will learn how to design the bioinformatics analysis of high throughput experiments. Given the background from the three earlier methods they should be able to design a pipeline that can easily be extended for more advanced analysis. After completing the course, the participants will understand the principles of experimental design and their relevance to high throughput experiments. The participants can design experiments and critically evaluate the influence of various design parameters. Examination elements are: pre-course individual assignment, and individual final presentation. For the first week, the PhD students will get pre-assignment tasks. The students can work on these pre-assignments at their home institutions. For the second week, all students will come to Stockholm to participate at the course on-site for the whole week. Past editions:
- In-person, 3 – 7 September, 2018 at SciLife Lab, Solna.
- In-person, 2 – 6 September, 2019 at SciLife Lab, Solna.
- In-person, 25 – 29 October, 2021 at SciLife Lab, Solna.
- Online, 19 – 23 September, 2022 and in-person at SciLife Lab, 26 – 30 September, 2022.
- In-person, 13 – 17 November, 2023 at SciLife Lab, Solna.
- In-person, 3-7 November 2025 at SciLifeLab Solna
From 2026 onwards, MedBioInfo students will take this course together with DDLS-RS students.
Second- or Third-year courses
Computational Systems Biology, 3-5 HP (Mika Gustafsson, Linköping Univ). Students will study basic systems biology and its application in biomedical research and medical problems. The course introduces basic concepts and mathematics underlying modern systems biology. You will understand the differences between small-scale and large-scale models using examples from biomedical research. The course will emphasise large-scale systems biology methods. The standard course provides 3HP, but there is an option to complete further assignments for 5HP. This course will run every other year, and all second- and third-year students will receive automatic invites.
In 2025, the course ran 5-8 May in Linköping.
2026-27 edition, dates TBA.
Third-year courses:
5. Career alternatives and your scientific CV, 1HP (Carsten Daub, Karolinska Inst.). In this course the students will consider career pathways in academia and industry. They will perform exercises on their CV and handling their interview. There will be a for-credit assignment for these purposes. A scientist employed in industry may join as instructor / coach.
Will run for the first time in 2026, dates TBA.
New course, 3-5 HP. Topic, teacher, and dates are under discussion and will be announced.
Taking courses in an alternative order due to parental leave or other reasons:
If you an enrolled student taking parental leave, first of all, congratulations, and enjoy! Secondly, naturally any courses you missed can be taken after you return if they are still offered. The caveat is that enrollments would not be automatic, rather you would be responsible for contacting the teacher to request admission.
If you wish to take any courses earlier than suggested, this is OK if you and the teacher feel you are ready. We don’t recommend delaying courses, because later in your PhD you will have less time for courses, and also less time to apply the training to your thesis. However there are many situations in which it makes sense to delay a course, and it is typically possible to accommodate you.
Meetings (1 hp)
Another part of the research school are the meetings.
The 2020 meeting was much shorter than usual and run by Zoom on 19 May. This was limited to student meeting, students reporting to the PIs, and a PI meeting. No certificates or HP were issued for that meeting.
Our first post-pandemic in-person meeting was at Grand Hotell Saltsjöbaden, 4-6 October 2021.
In 2022 we had two meetings, the first in Åre, 28-30 March. The second was jointly with NORBIS at Rosendal, Norway, 24-28 October 2022.
Our 2023 meeting will be at Yasuragi in the Stockholm Archipelago, 11-13 September. It will run jointly with NORBIS.
In addition to increasing the communication between the medical and informatics community the Meeting is also an important part of the education of the Ph.D. students. The Meeting under ordinary circumstances includes:
1. Training in soft skills.
2. Posters
3. Individual mentoring by the bioinformatics mentor.
4. Social activities
5. Research talks by students and PIs.
6. Unstructured time for discussions.
Some photos from the 2018 meeting are here.
Cancellations: If you sign up for one of our meetings but cancel after the deadline, you (meaning your Department) are liable for any cancellation costs, which can be up to 100% of the hotel fees plus certain other costs.
Completion certificate
We will provide a completion certificate to students that fullfill either of the following requirements:
- Four of the required courses and two Annual Meetings
- Five of the required courses and one Annual Meeting
The Studierektor can, upon student request, review a sufficiently-similar non-MedBioInfo course and determine whether it can be counted toward the above course requirement. The certificate outlines the school’s goals and obligatory elements, and describes the competitive acceptance criteria. This certificate is not a degree and does not itself confer any additional ECTS points.
Outside courses
There are of course many fine courses available outside the Research School, in Sweden, abroad, or online, which we encourage students to take as appropriate. Programming courses are important for first-year students who have not had them (or the equivalent experience). SciLifeLab offers courses — the Python may be particularly appropriate. Watch for the registration deadline for this programming course. Some of the core courses require programming, and students are responsible for meeting all prerequisites in time to take courses. Some advanced students may find other NBIS courses such as Single-Cell Analysis useful.
Mentoring
One of the aspects of this program is to improve the supervision of the students. Here, we propose to use a modified version of the idea used in Gothenburg some time ago with two supervisors for each student, one bioinformatician and one medical scientist. Our goal is that this program each student will be assigned a mentor from the Swedish bioinformatics community. The mentor should be from another research constellation than the student. The role of the mentor will be to help the student identify and solve computational bottlenecks in their research project, and also to provide more general career advice. The student will discuss with the mentor at least twice per year, either via electronic meetings or by short visits. We suggest using this Student-Mentor-meeting-form to guide student-mentor discussions.